Method development in the competence area Bioinformatics
Method development in the competence area Bioinformatics
Several projects on methodological developments have emerged from our research approach to molecularly explainable intelligence. The developments range from deep learning approaches for the precise localization of disease patterns to geometric optimization procedures for maximizing sample yield in laser capture microdissection. Our methodological developments also include software platforms for infrared microscopy.
Deep Learning approaches for the precise localization of disease patterns
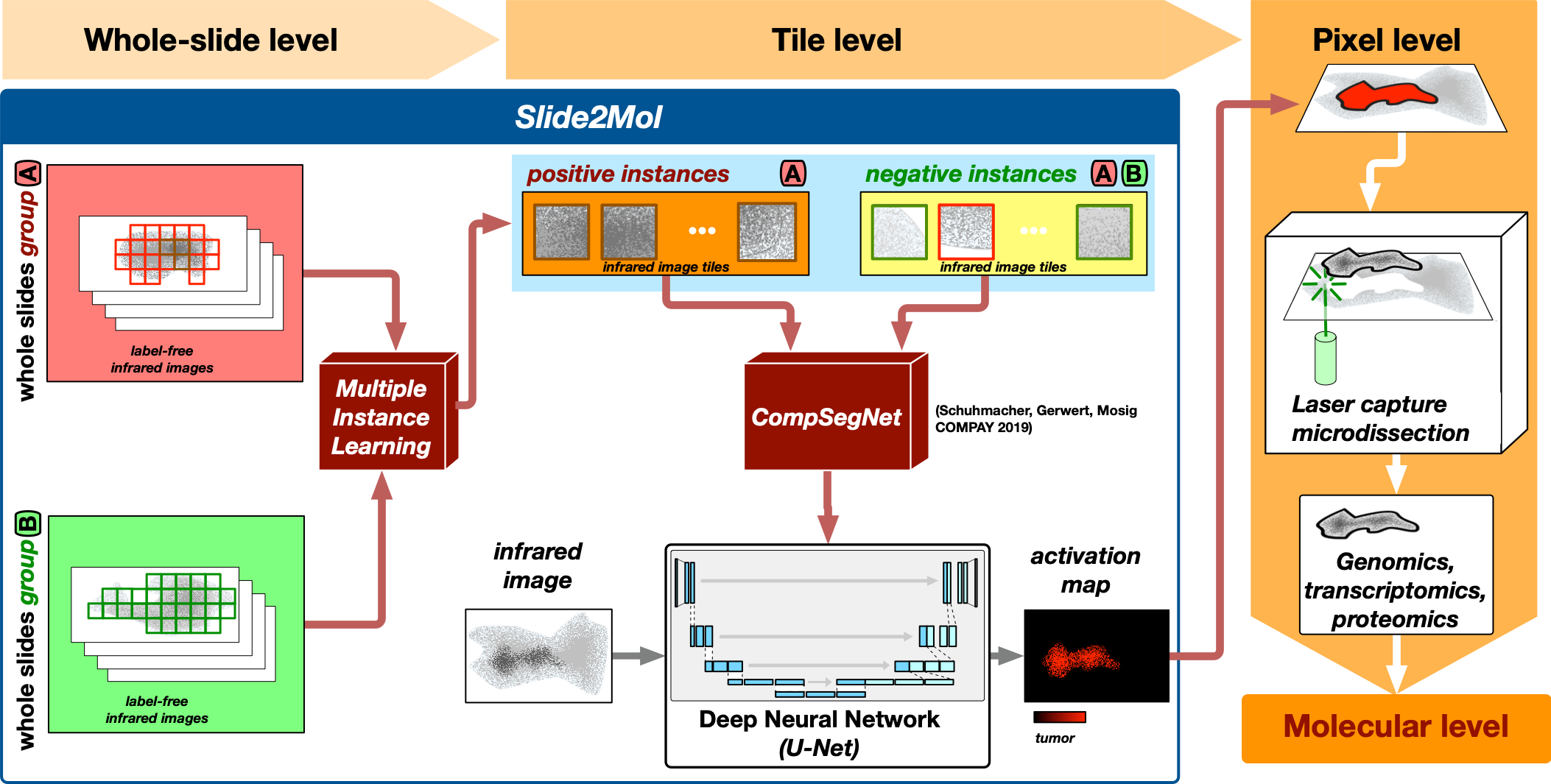
Microscopic imaging of tissue thin sections yields enormous amounts of data. A single infrared microscopic image of a tissue sample comprises hundreds of millions of infrared spectra, each providing a highly location specific molecular fingerprint of the sample. In bioinformatics we deal with the problem of localizing the disease-specific regions from this enormous amount of location specific spectral information. To do this, we use various deep learning approaches, including multiple instance learning and other so-called weakly supervised learning approaches, in particular our in-house developed comparative segmentation network.
Geometric optimisation for laser capture microdissection
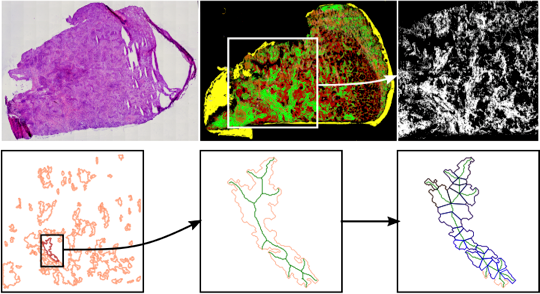
Laser capture microdissection (LMD) makes it possible to cut out specific areas from tissue thin-sections with high precision and to characterize them molecularly. This makes the LMD an ideal companion technology to the label-free infrared microscopy established in PRODI. However, in order to be able to use LMD optimally in the context of clinical studies, the tissues to be extracted must be broken down into fractional shapes according to certain geometric criteria. For this purpose, we have developed appropriate geometric optimization methods in cooperation with Prof. Maike Buchin (Faculty of Computer Science, RUB).
Software platforms for infrared microscopy

In bioinformatics, software platforms are a mainstay of scientific infrastructure. Open scientific software ensures reproducibility and enables different approaches to be compared. Software platforms promote the establishment of new data-driven technologies and accelerate the transfer. To foster this in the field of infrared microscopy, the methodical contributions from the competence area bioinformatics are made available as part of the DFG-funded OpenVibSpec platform and thus contribute to the further development of infrared microscopy-based label-free digital pathology.