Bioinformatics
Research focus
Artificial Intelligence for Protein Diagnostics
Artificial intelligence (AI) is an essential approach to fully exploit the potential of the molecular and analytical methods established in PRODI in protein diagnostics. The signatures of disease-specific molecular changes are often present in biomedical data in masked form and cannot be used directly for diagnostic purposes. This is where artificial intelligence comes into play: If sufficiently large data sets are collected in clinical studies, the decisive patterns can be uncovered using techniques such as so-called deep learning and used, for example, to distinguish between subtypes of cancer. Accordingly, the research focus in the bioinformatics competence area is on establishing AI techniques to uncover disease patterns and use them diagnostically.
Method development
Molecularly Interpretable Artificial Intelligence
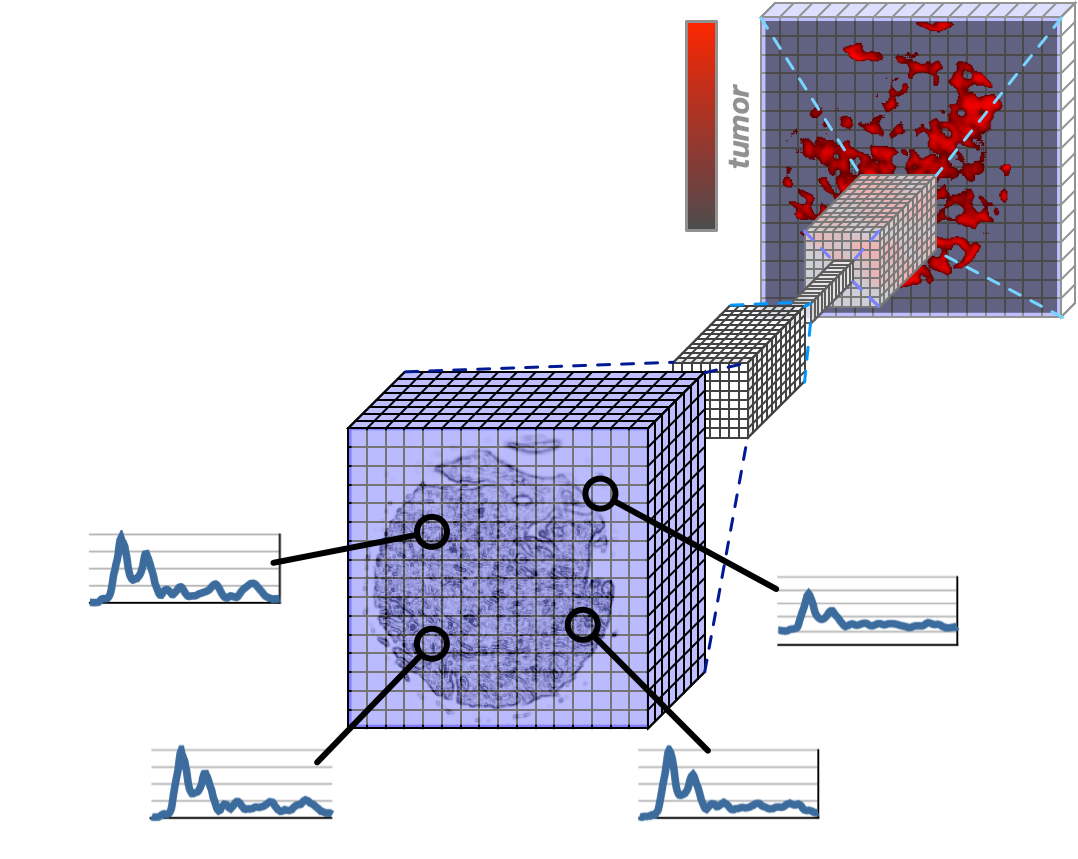
The focus of our method development is on questions of interpretable artificial intelligence: When an AI system makes a diagnostic classification of a sample, initially neither patients nor doctors can understand how the AI system arrived at this decision. Our research approach addresses this problem by developing algorithms for the molecular interpretability of AI-based decisions. In addition to the actual classification of the disease status, a molecular interpretation consists of an additional, extended prediction of a molecular property of the classified sample. The advantage of this approach is that the predicted molecular property can be experimentally tested on each individual sample from each individual patient. If the extended prediction of the molecular property is correct, the classification of the disease status is also trustworthy.
Such molecularly interpretable AI systems require the development of new algorithms and approaches. In the bioinformatics competence area in ProDi, we are developing such methods on the basis of deep neural networks, i.e. so-called deep learning.